Select furanose type:
Paste PDB data -> |
|
Note: Error reports are welcome (Semtember, 2021).
Sample data 1 (Standard)
HETATM 3632 C1 BXX A 602 20.048 19.283 5.331 1.00 32.19 C
HETATM 3639 C2 BXX A 602 19.621 18.280 4.247 1.00 30.41 C
HETATM 3637 C3 BXX A 602 18.523 19.023 3.508 1.00 31.64 C
HETATM 3634 C4 BXX A 602 18.885 20.497 3.706 1.00 30.78 C
HETATM 3633 O4 BXX A 602 19.404 20.538 5.023 1.00 32.68 O
HETATM 3631 O1 BXX A 602 19.720 18.886 6.687 1.00 34.55 O
HETATM 3635 C5 BXX A 602 17.763 21.513 3.515 1.00 30.88 C
HETATM 3636 O5 BXX A 602 16.688 21.216 4.401 1.00 28.49 O
HETATM 3638 O3 BXX A 602 18.416 18.667 2.097 1.00 26.17 O
HETATM 3640 O2 BXX A 602 19.296 16.928 4.714 1.00 25.23 O
Sample data 2 (Fructose)
HETATM 3639 C2 FRU A 702 20.290 19.543 5.543 1.00 25.30 C
HETATM 3636 C3 FRU A 702 20.076 18.440 4.492 1.00 22.03 C
HETATM 3634 C4 FRU A 702 18.960 19.071 3.628 1.00 21.36 C
HETATM 3633 C5 FRU A 702 19.094 20.577 3.834 1.00 21.41 C
HETATM 3638 O5 FRU A 702 19.405 20.619 5.184 1.00 22.23 O
HETATM 3631 O6 FRU A 702 16.779 21.110 4.404 1.00 20.39 O
HETATM 3632 C6 FRU A 702 17.891 21.458 3.553 1.00 21.24 C
HETATM 3635 O4 FRU A 702 18.915 18.625 2.220 1.00 19.36 O
HETATM 3637 O3 FRU A 702 19.723 17.060 4.944 1.00 19.69 O
HETATM 3640 O2 FRU A 702 20.089 19.046 6.903 1.00 23.16 O
HETATM 3641 C1 FRU A 702 21.659 20.281 5.378 1.00 27.10 C
HETATM 3642 O1 FRU A 702 22.737 19.429 5.694 1.00 29.49 O
Sample data 3 (DNA)
ATOM 2129 O5' DT N 1 -6.208 19.857 60.270 1.00 93.16 O
ANISOU 2129 O5' DT N 1 12950 11912 10532 -829 2239 -113 O
ATOM 2130 C5' DT N 1 -7.045 21.005 60.455 1.00 94.56 C
ANISOU 2130 C5' DT N 1 12944 12325 10659 -601 2291 -124 C
ATOM 2131 C4' DT N 1 -6.454 22.019 61.428 1.00 93.35 C
ANISOU 2131 C4' DT N 1 13035 11887 10544 -435 2285 9 C
ATOM 2132 O4' DT N 1 -6.137 21.394 62.702 1.00 92.77 O
ANISOU 2132 O4' DT N 1 13231 11548 10468 -668 2343 6 O
ATOM 2133 C3' DT N 1 -5.132 22.665 61.028 1.00 91.45 C
ANISOU 2133 C3' DT N 1 12975 11371 10398 -258 2159 159 C
ATOM 2134 O3' DT N 1 -5.328 23.666 60.024 1.00 92.03 O
ANISOU 2134 O3' DT N 1 12903 11606 10456 40 2132 205 O
ATOM 2135 C2' DT N 1 -4.734 23.268 62.371 1.00 90.86 C
ANISOU 2135 C2' DT N 1 13153 11041 10329 -249 2205 218 C
ATOM 2136 C1' DT N 1 -5.140 22.167 63.359 1.00 91.31 C
ANISOU 2136 C1' DT N 1 13293 11061 10337 -534 2285 133 C
ATOM 2137 N1 DT N 1 -3.995 21.282 63.846 1.00 89.91 N
ANISOU 2137 N1 DT N 1 13400 10574 10186 -688 2214 172 N
ATOM 2138 C2 DT N 1 -2.965 21.810 64.626 1.00 88.97 C
ANISOU 2138 C2 DT N 1 13510 10202 10089 -610 2156 251 C
ATOM 2139 O2 DT N 1 -2.885 22.983 64.969 1.00 88.78 O
ANISOU 2139 O2 DT N 1 13504 10146 10080 -462 2176 292 O
ATOM 2140 N3 DT N 1 -1.990 20.906 64.998 1.00 88.14 N
ANISOU 2140 N3 DT N 1 13630 9890 9966 -702 2078 264 N
ATOM 2141 C4 DT N 1 -1.933 19.554 64.691 1.00 88.57 C
ANISOU 2141 C4 DT N 1 13762 9909 9979 -852 2070 228 C
ATOM 2142 O4 DT N 1 -1.011 18.832 65.076 1.00 87.86 O
ANISOU 2142 O4 DT N 1 13913 9626 9841 -863 2000 251 O
ATOM 2143 C5 DT N 1 -3.031 19.064 63.883 1.00 89.53 C
ANISOU 2143 C5 DT N 1 13668 10257 10092 -978 2158 147 C
ATOM 2144 C7 DT N 1 -3.091 17.619 63.480 1.00 89.97 C
ANISOU 2144 C7 DT N 1 13821 10272 10089 -1185 2193 85 C
ATOM 2145 C6 DT N 1 -3.988 19.935 63.507 1.00 90.20 C
ANISOU 2145 C6 DT N 1 13474 10601 10195 -895 2215 114 C
Sample data 4 (NADH)
HETATM11465 PA NAI A 401 -32.220 -16.579 6.480 1.00 21.60 P
HETATM11466 O1A NAI A 401 -30.874 -16.161 5.916 1.00 22.93 O
HETATM11467 O2A NAI A 401 -32.770 -17.924 6.155 1.00 23.29 O
HETATM11468 O5B NAI A 401 -32.170 -16.364 8.047 1.00 22.56 O
HETATM11469 C5B NAI A 401 -33.301 -16.505 8.885 1.00 23.85 C
HETATM11470 C4B NAI A 401 -32.881 -16.983 10.262 1.00 21.56 C
HETATM11471 O4B NAI A 401 -34.076 -17.051 11.054 1.00 22.54 O
HETATM11472 C3B NAI A 401 -32.205 -18.340 10.280 1.00 21.39 C
HETATM11473 O3B NAI A 401 -30.851 -18.199 10.715 1.00 20.03 O
HETATM11474 C2B NAI A 401 -33.046 -19.145 11.241 1.00 22.62 C
HETATM11475 O2B NAI A 401 -32.323 -20.027 12.115 1.00 23.85 O
HETATM11476 C1B NAI A 401 -33.769 -18.045 12.015 1.00 22.36 C
HETATM11477 N9A NAI A 401 -34.951 -18.594 12.674 1.00 21.39 N
HETATM11478 C8A NAI A 401 -35.825 -19.493 12.181 1.00 23.94 C
HETATM11479 N7A NAI A 401 -36.747 -19.794 13.117 1.00 24.54 N
HETATM11480 C5A NAI A 401 -36.476 -19.056 14.209 1.00 22.64 C
HETATM11481 C6A NAI A 401 -37.021 -18.884 15.572 1.00 22.90 C
HETATM11482 N6A NAI A 401 -38.121 -19.536 15.956 1.00 22.37 N
HETATM11483 N1A NAI A 401 -36.405 -18.031 16.427 1.00 22.75 N
HETATM11484 C2A NAI A 401 -35.319 -17.336 16.062 1.00 24.15 C
HETATM11485 N3A NAI A 401 -34.757 -17.436 14.836 1.00 22.97 N
HETATM11486 C4A NAI A 401 -35.281 -18.282 13.905 1.00 22.35 C
HETATM11487 O3 NAI A 401 -33.349 -15.510 6.011 1.00 19.91 O
HETATM11488 PN NAI A 401 -32.967 -13.988 5.619 1.00 19.27 P
HETATM11489 O1N NAI A 401 -32.107 -13.369 6.682 1.00 17.81 O
HETATM11490 O2N NAI A 401 -32.539 -13.943 4.164 1.00 19.26 O
HETATM11491 O5D NAI A 401 -34.477 -13.415 5.609 1.00 17.59 O
HETATM11492 C5D NAI A 401 -35.131 -13.085 6.851 1.00 17.27 C
HETATM11493 C4D NAI A 401 -36.259 -12.086 6.537 1.00 17.94 C
HETATM11494 O4D NAI A 401 -35.695 -10.969 5.857 1.00 20.00 O
HETATM11495 C3D NAI A 401 -37.298 -12.632 5.572 1.00 18.07 C
HETATM11496 O3D NAI A 401 -38.580 -12.213 5.999 1.00 16.78 O
HETATM11497 C2D NAI A 401 -36.909 -12.074 4.191 1.00 19.24 C
HETATM11498 O2D NAI A 401 -38.019 -11.859 3.340 1.00 18.10 O
HETATM11499 C1D NAI A 401 -36.271 -10.745 4.564 1.00 20.72 C
HETATM11500 N1N NAI A 401 -35.214 -10.256 3.724 1.00 23.44 N
HETATM11501 C2N NAI A 401 -35.129 -8.932 3.501 1.00 30.76 C
HETATM11502 C3N NAI A 401 -34.113 -8.334 2.694 1.00 31.80 C
HETATM11503 C7N NAI A 401 -34.069 -6.831 2.411 1.00 30.30 C
HETATM11504 O7N NAI A 401 -33.383 -6.329 1.494 1.00 27.00 O
HETATM11505 N7N NAI A 401 -34.809 -6.019 3.196 1.00 29.60 N
HETATM11506 C4N NAI A 401 -33.144 -9.222 2.161 1.00 36.58 C
HETATM11507 C5N NAI A 401 -33.221 -10.606 2.465 1.00 34.36 C
HETATM11508 C6N NAI A 401 -34.263 -11.090 3.254 1.00 30.54 C
Input lines must follow the PDB guideline.
Do not change the COLUMNS of atom name (13-16), x (31-38), y (39-46), and z (47-54) coordinates.
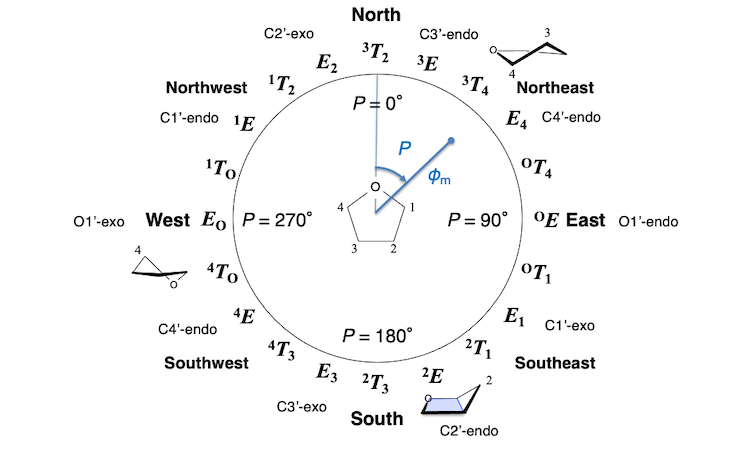
What's Altona-Sundaralingam parameter?
It is the puckering coordinate system of a five-membered furanose ring [1,2]. φm (or γm) means the magnitude of puckering. P means the phase angle. Ribose of nucleic acids frequently takes C2'-endo (2E, P ~ 160°) or C3'-endo (3E, P ~ 18°) conformation.
References
-
C. Altona & M. Sundaralingam: Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J. Am. Chem. Soc. 94, 8205-12 (1972) [doi]
-
H.A. Taha, M.R. Richards & T.L. Lowary: Conformational analysis of furanoside-containing mono- and oligosaccharides. Chem. Rev. 113, 1851-76 (2013) [doi]
About citation: No need to cite this URL because resources on The Internet are unstable. I would be happy if you acknowledge my name (Shinya Fushinobu) when you feel this site was helpful.